Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues [full article]
Taeyun Ku, Justin Swaney, Jeong-Yoon Park, Alexandre Albanese, Evan Murray, Jae Hun Cho, Young-Gyun Park, Vamsi Mangena, Jiapei Chen & Kwanghun Chung (2016) Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nature Biotechnology 34:973–981.
We highly recommend eMAP instead of MAP. [eMap Protocol] (A detailed instruction page will be made available soon.)
Introduction
MAP (Magnified Analysis of Proteome) is a tissue processing technology that transforms intact biological tissues into transparent, size-adjustable tissues. Using MAP, tissue is hybridized with a unique hydrogel—dense polyacrylamide hydrogel with polyelectrolyte, which preserves target macrobiomolecules such as proteins effectively. The size of the tissue-hydrogel hybrid can be adjusted depending on its mounting media. MAP tissue can be expanded about fourfold linearly which allows super-resolution imaging using conventional light microscopes such as confocal laser scanning microscope. The tissue can be also shrunk back to the original size to accommodate fast whole-tissue imaging. This size-adjustable property is reversible and repeatable, which enables multiscale imaging of the same sample, offering both organ-level protein localization and fine 3D visualization at the nanoscale.
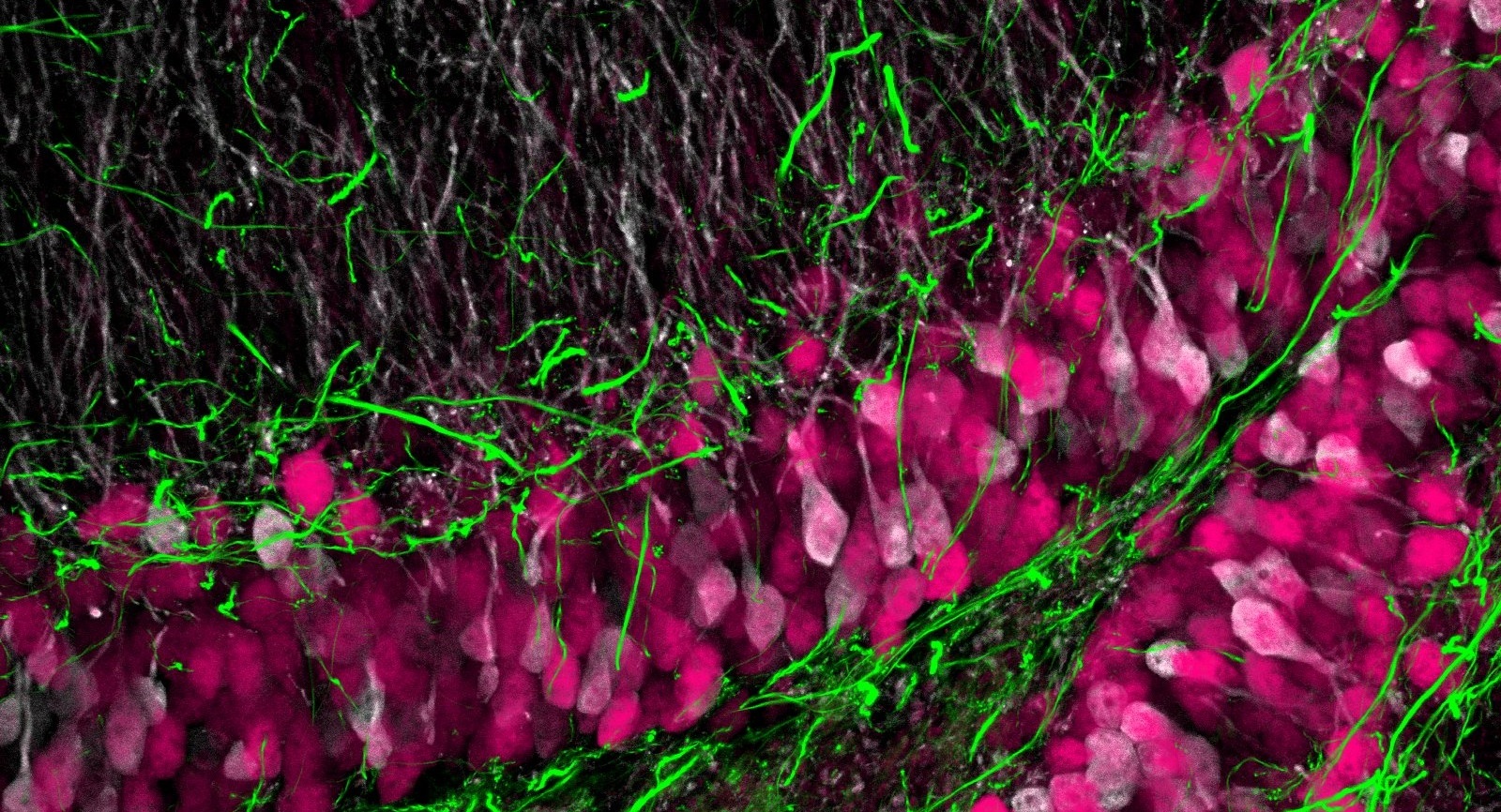
MAP reveals 3D nanoscale structures in biological tissues such as cytoskeleton, synapses, and neurofilaments. As such, MAP enables isolating pre- and post-synaptic structures, and improves the accuracy of neural fiber tracing.
Due to the preservation of macrobiomolecules in the robust tissue-hydrogel hybrid, MAP enables multiplexed immunolabeling of proteins via repeated antibody labeling and elution. We have demonstrated seven rounds of multiplexing with diverse cell-type and neurofilament markers in the report.
With CLARITY-optimized objectives having long working distances, MAP allows super-resolution imaging of millimeters-deep regions. We prepared a MAP-processed 2-mm-thick mouse brain slice (8 mm after expansion) and imaged the whole thickness at a consistent signal level as shown in the figure below. Our advanced labeling technologies based on stochastic electrotransport aids deep-tissue, large-scale super-resolution imaging.